MK
: Exact Algorithm to
Find Time
Series Motifs
This is a supporting page to our paper - Exact
Discovery of Time Series
Motifs, by Abdullah Mueen, Eamonn Keogh, Qiang Zhu,
Sydney Cash and Brandon Westover.
The paper is here.
This ppt contains some
additional information about the algorithm and the experimets.
|
| Codes
and Executables |
We
have two
versions of MK;
one for general time series motif and the other for
subsequence
motif. We also have the codes for the Brute Force motif finding
algorithm which are lot slower than MK. Click here to
download
the codes. Codes are written in C and compiled in windows
environment. Please follow the instructions in the Readme.txt
in each subfolders for detailed information..
|
| Randomwalk
Generator |
In the paper we used
several large randomwalk datasets which are too large for downloads. So
we provide the MATLAB code of the random
walk generator and the seeds for reproducing
the datasets exactly. For example, if the seed is
[123456 , 987654], a set of 10,000 random walks of length 1024 can be
generated by the following MATLAB code. The dataset will be saved in
ascii format in the 't.txt' file.
s = [123456 ;
987654];
randn('state',s);
c =
random_walk(1024,10000);
save('t.txt','c','-ascii');
The seeds for a particular experiment can be found in subsequent
sections in this page.
|
| Spreadsheet
of Experimental Resluts |
We
have compiled
results of all the experiments in an excel
document. In the subsequent sections, we will refer to sheets
from this excel document. The numbers in the spreadsheet can be
reproduced
exactly at every run. Execution times may not be exactly the same as in
the spreadsheet because of the random referencing, but they should be
very close and representative for the claim of our paper. It
is worth mentioning some notes about the document.
- All the real datasets are single time series and
processed by the definition of subsequence
motif using the mk_s
code.
- Locations of motifs are all indexed from 0 as in
standard C proframming language.
- All the times are in seconds.
- All the distances are distances in z-normalized space.
- All red values are extrapolated from the previous
experimental values.
- For Brute Force without
Early Abandonning the we extrapolate time in proportion of m^2.
- For Brute Force with
Early Abandonning the we extrapolate time in proportion of around m^1.8.
|
| Performance
Comparison |
Figure 5: The seeds
for the 10 random walk datasets are here.
All the execution times for the 10 runs on each of these 10 datasets
can be found in the "speedup details" sheet. Execution times for the
Brute Force algorithms can be found in the "speedup summary" sheet.
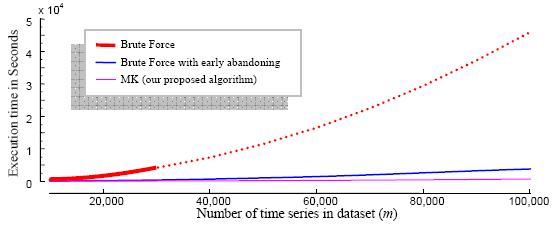 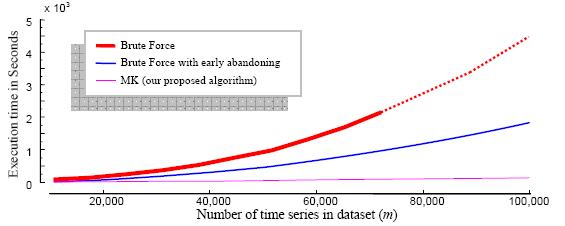
Figure 6: The
LSF5_10.txt
is a time series of length 180,214 which is a sampled eeg data. The datasets
for this experiment are sampled from the start of LSF5_10. All the
execution times for the 10 runs on each of the 10 datasets in it
can be found in the "speedup (real) details" sheet. Execution times for
the
Brute Force algorithms can be found in the "speedup (real) summary"
sheet.
|
| Why
is Early-Abandoning so Effective? |
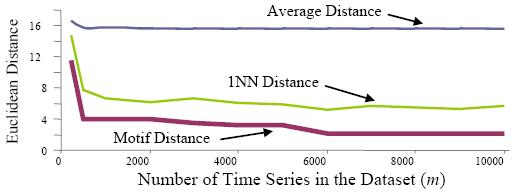
| Figure 7: We did
this experiment on small samples from the LSF5 dataset. The samples can
be found here. Statistics for
this experiments (motif distances,
average 1-NN distances and average distances) are found using BF_s code. |
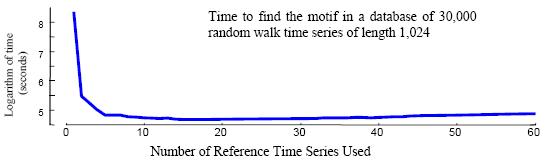
| Choosing
the Number of Reference Points |
Figure 8: We used
this random_shuffle function to shuffle the 30,000 random walks from
the Figure 5
dataset. We ran the mk_d.exe for 14 times on such shuffled data.
|
| Finding Repeated Insect Behaviors |
Figure 14: The dataset is here. Its length is 33,021.
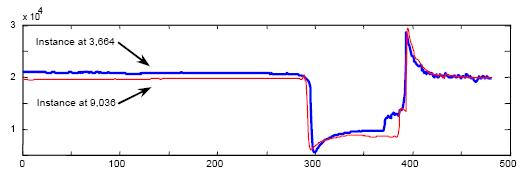 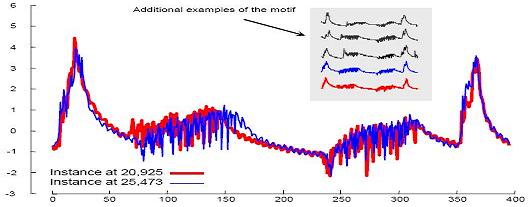
Errata: The data set for Figure:15 is of length 18,667 and can be found here.
The entire trace containing the above dataset is of length 78,254 and can be found here. |
| Near Duplication Detection |
The dataset we used have been taken from this page.
We used the dataset of 1.5 million tiny images and use this code to
generate 100,018 time series of length 768. These time series
are created from the first 100,018 images in the DFS order. Since the data file is very large, we give the code only.
|
| Automatically constructing EEG Dictionaries |
Figure 19 : The dataset for this experiment is LSF5_10.txt.
|
| Motif-Based
Anytime Time Series Classification |
| Datasets can be found here. |
|
This page is created by -
Abdullah Mueen
Department of Computer Science and Engineering,
University of California - Riverside. |